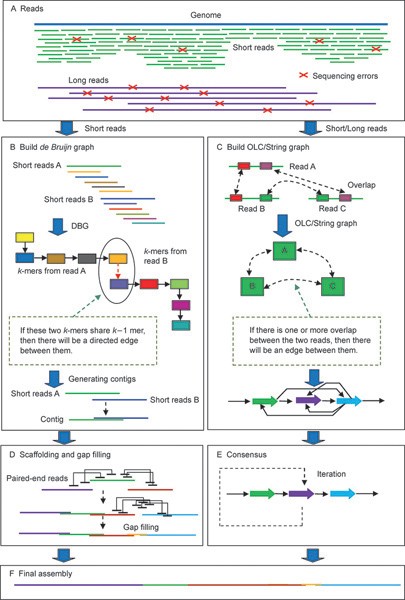
Flow chart of our reference guided genome assembly pipeline. All reads... | Download Scientific Diagram

High-throughput pipeline for the de novo viral genome assembly and the identification of minority variants from Next-Generation Sequencing of residual diagnostic samples | bioRxiv
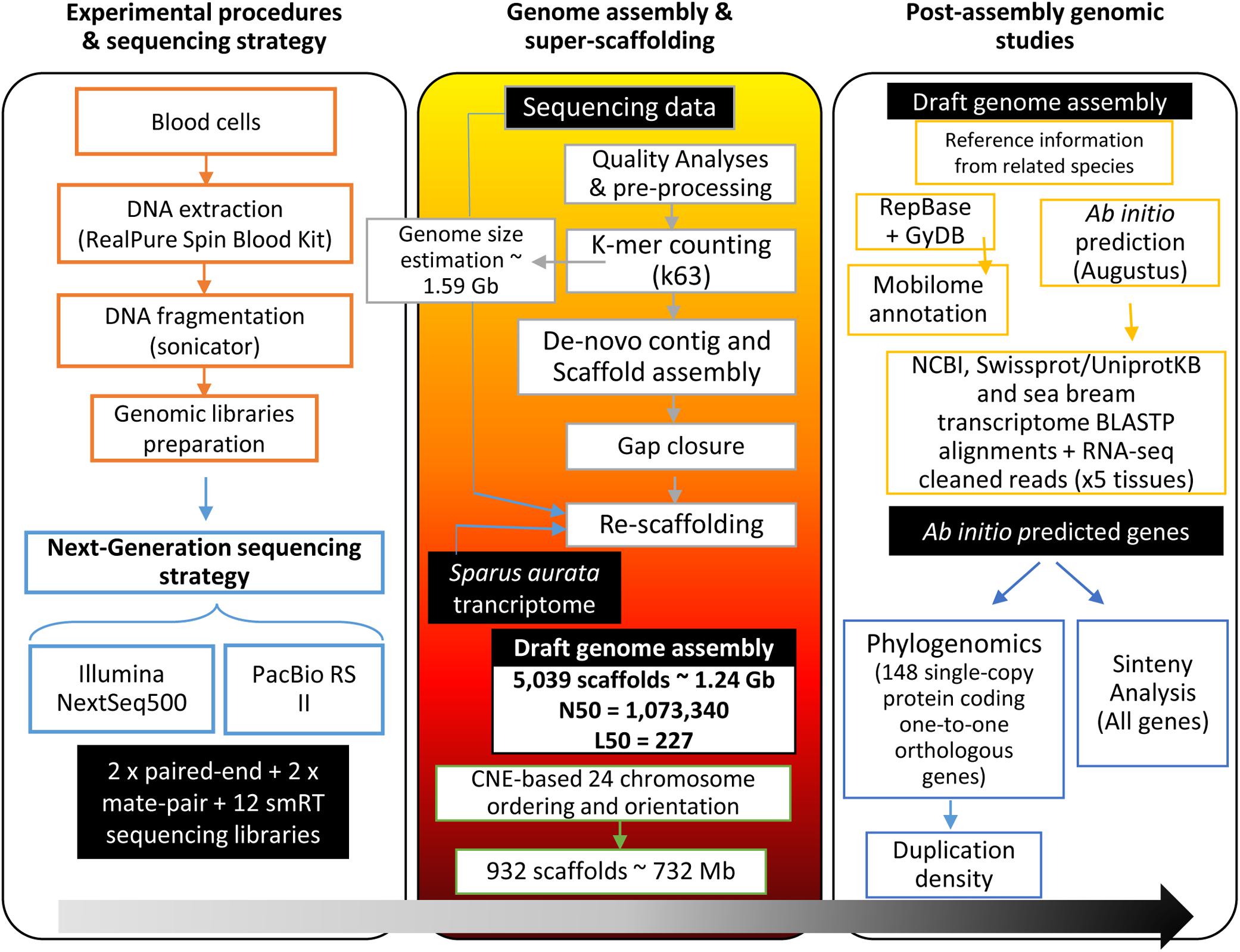
Frontiers | Genome Sequencing and Transcriptome Analysis Reveal Recent Species-Specific Gene Duplications in the Plastic Gilthead Sea Bream (Sparus aurata) | Marine Science
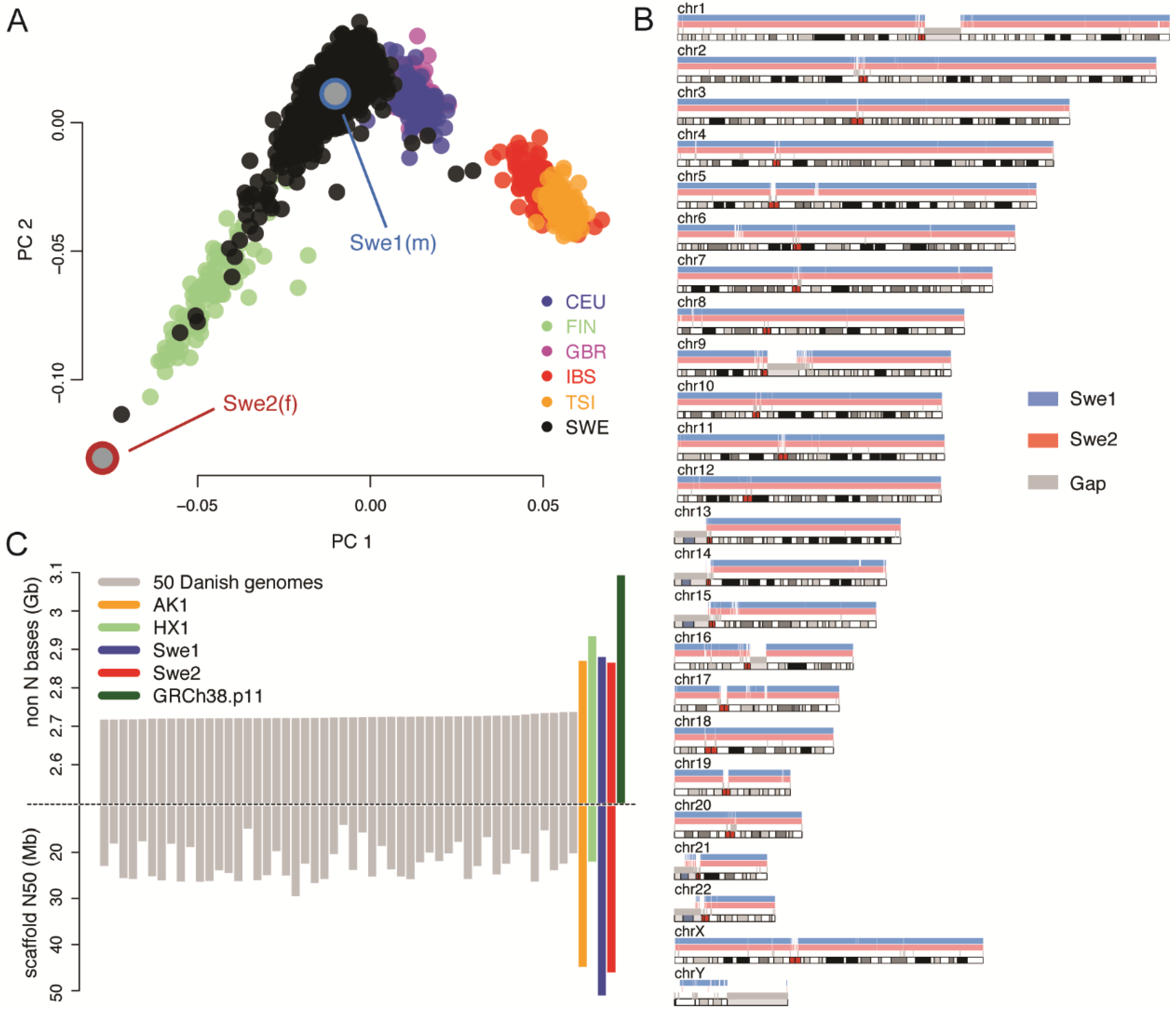
Genes | Free Full-Text | De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data | HTML

A comparative analysis of methods for de novo assembly of hymenopteran genomes using either haploid or diploid samples | Scientific Reports
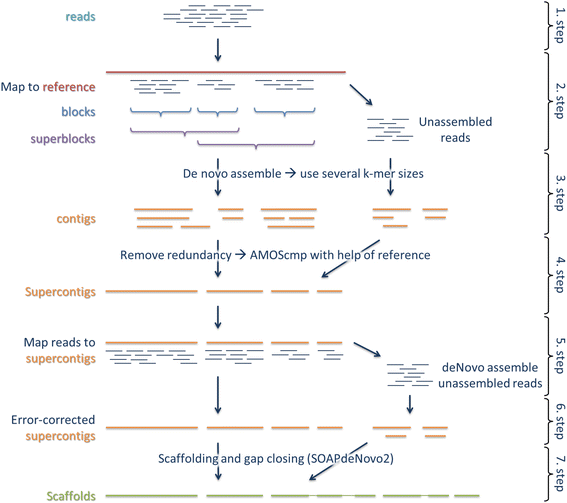
Reference-guided de novo assembly approach improves genome reconstruction for related species | BMC Bioinformatics | Full Text

De novo assembly of Oxford Nanopore MinION reads with QIAGEN CLC Genomics Workbench - Bioinformatics Software and Services | QIAGEN Digital Insights

Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions | Nature Communications

Closing Human Reference Genome Gaps: Identifying and Characterizing Gap-Closing Sequences | G3: Genes | Genomes | Genetics

Whole Genome de novo Assembly |Supporting de novo assembly training |Course |WGS sequencing | data analysis| Workshop